We talked about features selection based on Lasso(https://charleshsliao.wordpress.com/2017/04/11/regularization-in-neural-network-with-mnist-and-deepnet-of-r/), and autoencoder. More features will make the model more complex. it can be a good idea to reduce the number of features to only the most useful ones, and discard the rest. There are three basic strategies: Univariate statistics, model-based selection and iterative selection.
We use the data from sklearn library(need to download face datasets separately), and the IDE is sublime text3. Most of the code comes from the book: https://www.goodreads.com/book/show/32439431-introduction-to-machine-learning-with-python?from_search=true
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
###1. Univariate Statistics
###We add noise to our cancer dataset
from sklearn.datasets import load_breast_cancer
from sklearn.feature_selection import SelectPercentile
cancer=load_breast_cancer()
rng=np.random.RandomState(66)
noise=rng.normal(size=(len(cancer.data),50))
X_w_noise=np.hstack([cancer.data,noise])
###select 10% of features and transform training data
X_train, X_test, y_train, y_test = train_test_split(
X_w_noise, cancer.target, random_state=0, test_size=.5)
select=SelectPercentile(percentile=50)
select.fit(X_train,y_train)
X_train_selected=select.transform(X_train)
print(X_train.shape)
###(284, 80)
print(X_train_selected.shape)
###(284, 40)
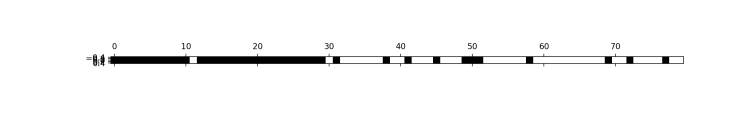
###We can find out which features have been selected using the get_support method,
###which returns a boolean mask of the selected features, with black as selected ones
s_features=select.get_support()
print(s_features)
plt.matshow(s_features.reshape(1, -1), cmap='gray_r')
plt.show()
###[ True True True True True True True True True False True False
### True True True True True True False False True True True True
### True True True True True True False False False True False True
### False False True False False False False True False False True False
### False True False True False False False False False False True False
### True False False False False True False True False False False False
### True True False True False False False False]
###Now let us fit a model with selected features
from sklearn.linear_model import LogisticRegression
X_test_selected=select.transform(X_test)
lr=LogisticRegression()
lr.fit(X_train, y_train)
print("Score with all features: %f"%lr.score(X_test,y_test))
###0.901754
lr.fit(X_train_selected,y_train)
print("Score with selected features: %f"%lr.score(X_test_selected,y_test))
###0.933333
###In this case, removing the noise features improved performance
###2. Model-based feature selection
###Decision trees and decision tree based models provide feature importances; Linear models
###have coefficients which can be used by considering the absolute value. Linear models with
###L1 penalty learn sparse coefficients, which only use a small subset of features.
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import RandomForestClassifier
select=SelectFromModel(RandomForestClassifier(n_estimators=100, random_state=42)
,threshold="median")
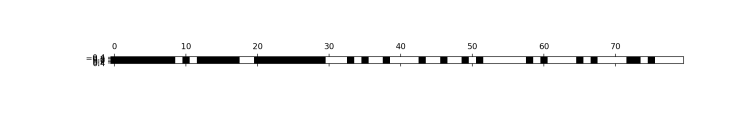
###The SelectFromModel class selects all features with importance measure of the feature higher than threshold
select.fit(X_train,y_train)
X_train_l1=select.transform(X_train)
print(X_train.shape)
#(284, 80)
print(X_train_l1.shape)
#(284, 40)
mask = select.get_support()
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.show()
X_test_l1 = select.transform(X_test)
print(LogisticRegression().fit(X_train_l1, y_train).score(X_test_l1, y_test))
###0.950877192982
###3.Iterative feature selection
###In iterative feature selection, a series of models is built, with varying numbers of
###features. There are two basic methods: starting with no features and adding features
###one by one, until some stopping criterion is reached, or starting with all features
###and removing features one by one, until some stopping criterion is reached.One particular
###method of this kind is recursive feature elimination (RFE)
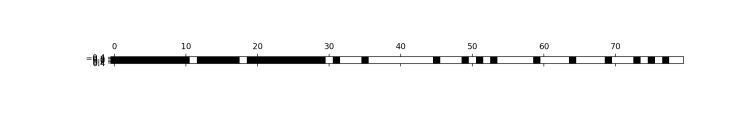
from sklearn.feature_selection import RFE
select = RFE(RandomForestClassifier(n_estimators=100, random_state=40),
n_features_to_select=40)
select.fit(X_train, y_train)
X_train_rfe=select.transform(X_train)
X_test_rfe=select.transform(X_test)
print(X_train_rfe.shape)
###(284, 40)
mask = select.get_support()
plt.matshow(mask.reshape(1, -1), cmap='gray_r')
plt.show()
print(LogisticRegression().fit(X_train_rfe, y_train).score(X_test_rfe, y_test))
###0.929824561404